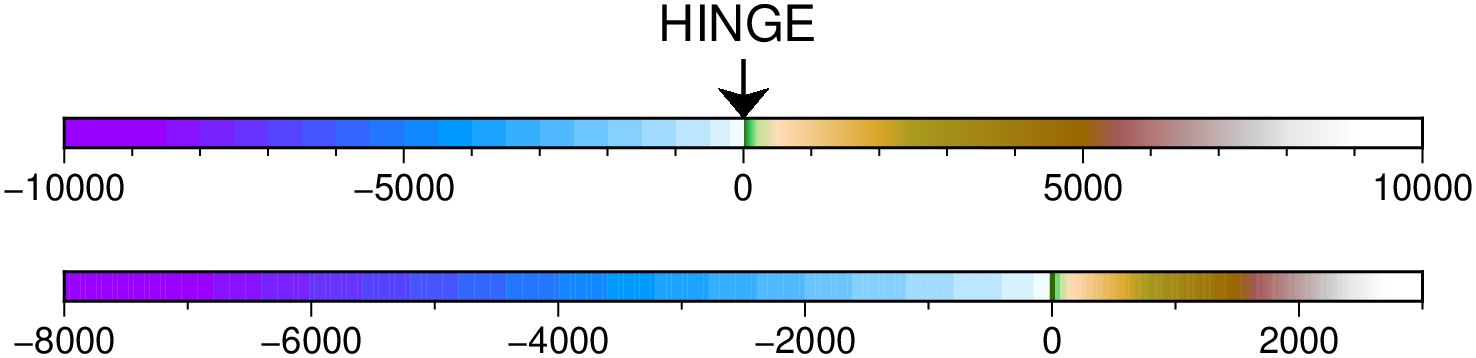
using GMT
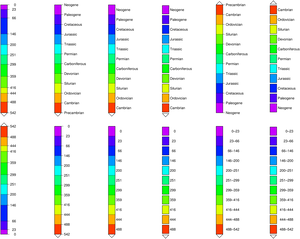
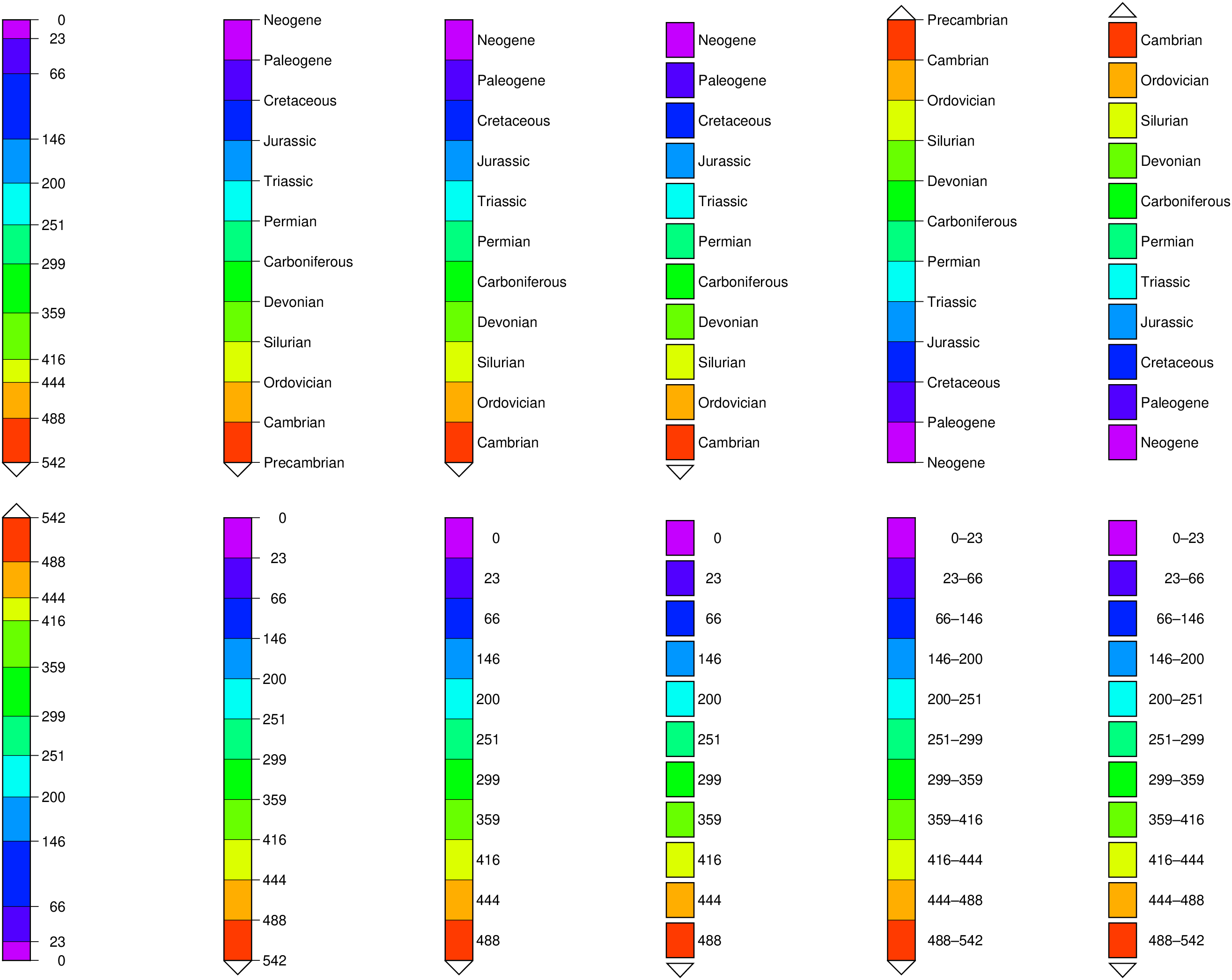
C = makecpt(cmap=((197,0,255),(81,0,255),(0,35,255),(0,151,255),(0,255,244),(0,255,127),(0,255,11),
(104,255,0),(220,255,0),(255,174,0),(255,58,0)),
T=[0,23,66,146,200,251,299,359,416,444,488,542]);
# Add the labels for the periods
C.label = ["Neogene", "Paleogene", "Cretaceous", "Jurassic", "Triassic", "Permian",
"Carboniferous", "Devonian", "Silurian", "Ordovician", "Cambrian;Precambrian"];
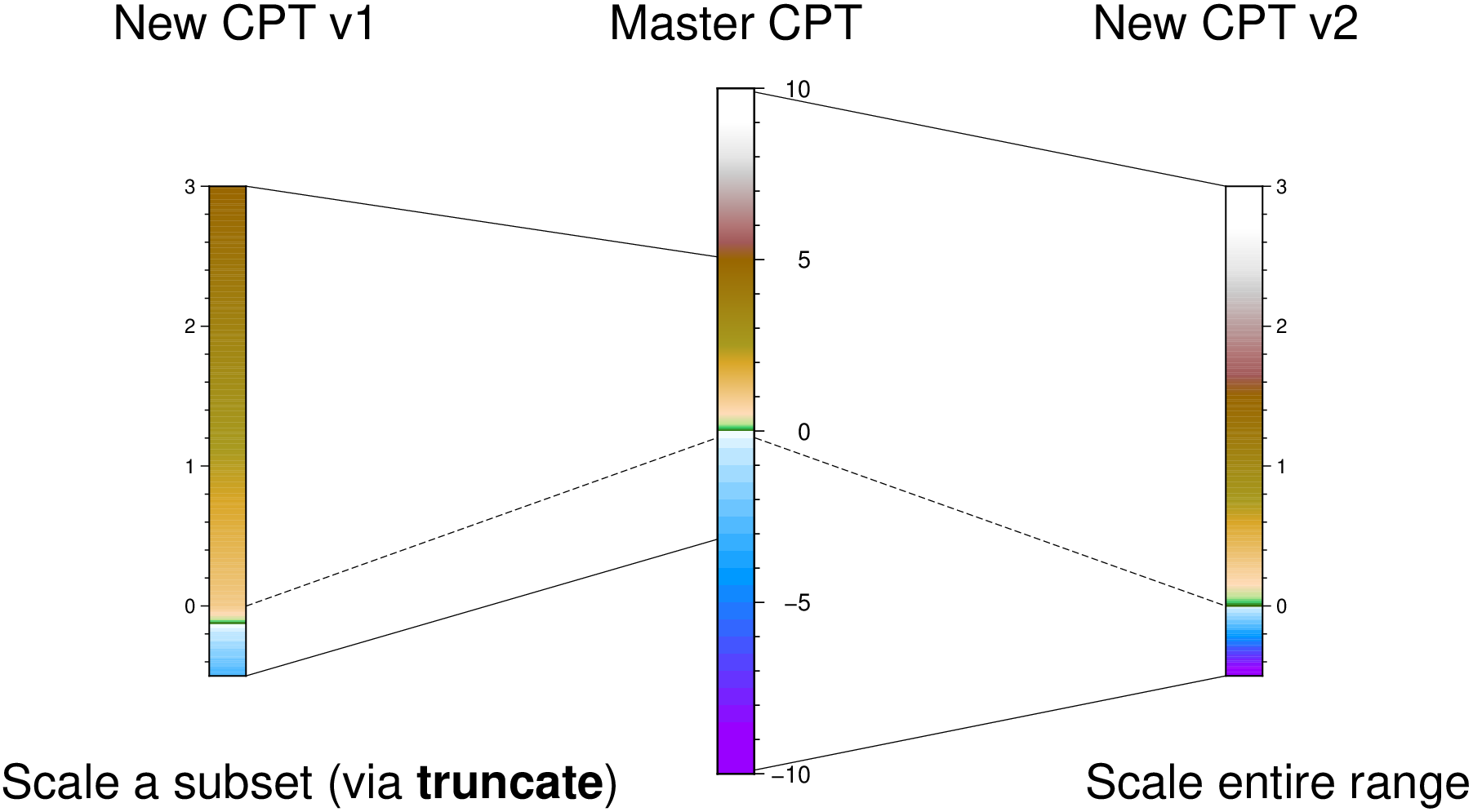
colorbar(pos=(paper=true, anchor=(0,13), size=(-8,0.5), justify=:ML, triangles=:f), B=:none)
colorbar!(pos=(paper=true, anchor=(4,13), size=(-8,0.5), justify=:ML, triangles=:f),
B=:none, equal_size=true)
colorbar!(pos=(paper=true, anchor=(8,13), size=(-8,0.5), justify=:ML, triangles=:f),
B=:none, equal_size=(gap=0,))
colorbar!(pos=(paper=true, anchor=(12,13), size=(-8,0.5), justify=:ML, triangles=:f),
B=:none, equal_size=(gap=0.1,))
colorbar!(pos=(paper=true, anchor=(16,13), size=(08,0.5), justify=:ML, triangles=:f),
B=:none, equal_size=true)
colorbar!(pos=(paper=true, anchor=(20,13), size=(08,0.5), justify=:ML, triangles=:f),
B=:none, equal_size=(gap=0.1,))
# Remove the labels so that we can plot the ages
C.label = fill("", length(C.label));
colorbar!(pos=(paper=true, anchor=(0,4), size=(08,0.5), justify=:ML, triangles=:f), B=:none)
colorbar!(pos=(paper=true, anchor=(4,4), size=(-8,0.5), justify=:ML, triangles=:f),
B=:none, equal_size=true)
colorbar!(pos=(paper=true, anchor=(8,4), size=(-8,0.5), justify=:ML, triangles=:f),
B=:none, equal_size=(gap=0,))
colorbar!(pos=(paper=true, anchor=(12,4), size=(-8,0.5), justify=:ML, triangles=:f),
B=:none, equal_size=(gap=0.1,))
colorbar!(pos=(paper=true, anchor=(16,4), size=(-8,0.5), justify=:ML, triangles=:f),
B=:none, equal_size=(range=true,))
colorbar!(pos=(paper=true, anchor=(20,4), size=(-8,0.5), justify=:ML, triangles=:f),
B=:none, equal_size=(range=true, gap=0.1), show=true)