LiDAR Point clouds to 3D surfaces ✨➡️🏔️
Contents
LiDAR Point clouds to 3D surfaces ✨➡️🏔️¶
In this tutorial, let’s use PyGMT to perform a more advanced geoprocessing workflow 😎
Specifically, we’ll learn to filter and interpolate a LiDAR point cloud onto a regular grid surface 🏁
At the end, we’ll also make a 🚠 3D perspective plot for the Digital Surface Model (DSM) produced through this exercise!
0️⃣ The LiDAR data¶
Let’s visit Wellington which is the capital city of New Zealand 🇳🇿.
They recently had a LiDAR survey done between March 2019 to March 2020 🛩️, with the point cloud and derived products published under an open CC BY 4.0 license.
OpenTopography link: https://doi.org/10.5069/G9K935QX
Bulk download location: https://opentopography.s3.sdsc.edu/minio/pc-bulk/NZ19_Wellington
Official 1m DSM from LINZ: https://data.linz.govt.nz/layer/105024-wellington-city-lidar-1m-dsm-2019-2020
🔖 References:
https://github.com/GenericMappingTools/foss4g2019oceania/blob/v1/3_lidar_to_surface.ipynb
https://github.com/weiji14/30DayMapChallenge2021/blob/v0.3.1/day17_land.py
First though, we’ll need to download a sample file to play with 🎮
Luckily for us, there is a function called pygmt.which that has to ability to do just that!
The download=True option can be used to download ⬇️ a remote file to our local working directory.
# Download LiDAR LAZ file from a URL
lazfile = pygmt.which(
fname="https://opentopography.s3.sdsc.edu/pc-bulk/NZ19_Wellington/CL2_BQ31_2019_1000_2138.laz",
download=True,
)
print(lazfile)
CL2_BQ31_2019_1000_2138.laz
Loading a point cloud 🌧️¶
Now we can use laspy to read in our example LAZ file into a pandas.DataFrame.
The data will be kept in a variable called ‘df’ which stands for dataframe.
# Load LAZ data into a pandas DataFrame
lazdata = laspy.read(source=lazfile)
df = pd.DataFrame(
data={
"x": lazdata.x.scaled_array(),
"y": lazdata.y.scaled_array(),
"z": lazdata.z.scaled_array(),
"classification": lazdata.classification,
}
)
Quick data preview ⚡¶
Let’s get the bounding box 📦 region of our study area using pygmt.info.
# Get bounding box region
region = pygmt.info(data=df[["x", "y"]], spacing=1) # West, East, South, North
print(f"Data points covers region: {region}")
Data points covers region: [1749760. 1750240. 5426880. 5427600.]
Check how many 🗃️ data points (rows) are in the table and get some summary statistics printed out.
# Print summary statistics
print(f"Total of {len(df)} data points")
df.describe()
Total of 7322702 data points
| x | y | z | classification | |
|---|---|---|---|---|
| count | 7.322702e+06 | 7.322702e+06 | 7.322702e+06 | 7.322702e+06 |
| mean | 1.750009e+06 | 5.427144e+06 | 6.536329e+01 | 4.477295e+00 |
| std | 1.337038e+02 | 1.713325e+02 | 4.428189e+01 | 2.212308e+00 |
| min | 1.749760e+06 | 5.426880e+06 | -1.377360e+02 | 1.000000e+00 |
| 25% | 1.749896e+06 | 5.426997e+06 | 3.380700e+01 | 3.000000e+00 |
| 50% | 1.750012e+06 | 5.427125e+06 | 6.012200e+01 | 5.000000e+00 |
| 75% | 1.750121e+06 | 5.427280e+06 | 9.645000e+01 | 5.000000e+00 |
| max | 1.750240e+06 | 5.427600e+06 | 3.186180e+02 | 1.800000e+01 |
More than 7 million points, that’s a lot! 🤯
Let’s try to take a quick look of our LiDAR elevation points on a map 🗺️
PyGMT can plot these many points, but it might take a long time ⏳, so let’s do a quick filter by taking every n-th row from our dataframe table.
df_filtered = df.iloc[::20]
print(f"Filtered down to {len(df_filtered)} data points")
Filtered down to 366136 data points
Now we can visualize these quickly on a map with PyGMT 😀
# Plot XYZ points on a map
fig = pygmt.Figure()
fig.basemap(frame=True, region=region, projection="x1:5000")
pygmt.makecpt(cmap="bukavu", series=[df.z.min(), df.z.max()])
fig.plot(
x=df_filtered.x, y=df_filtered.y, color=df_filtered.z, style="c0.03c", cmap=True
)
fig.colorbar(position="JMR", frame=["af+lElevation", "y+lm"])
fig.show()
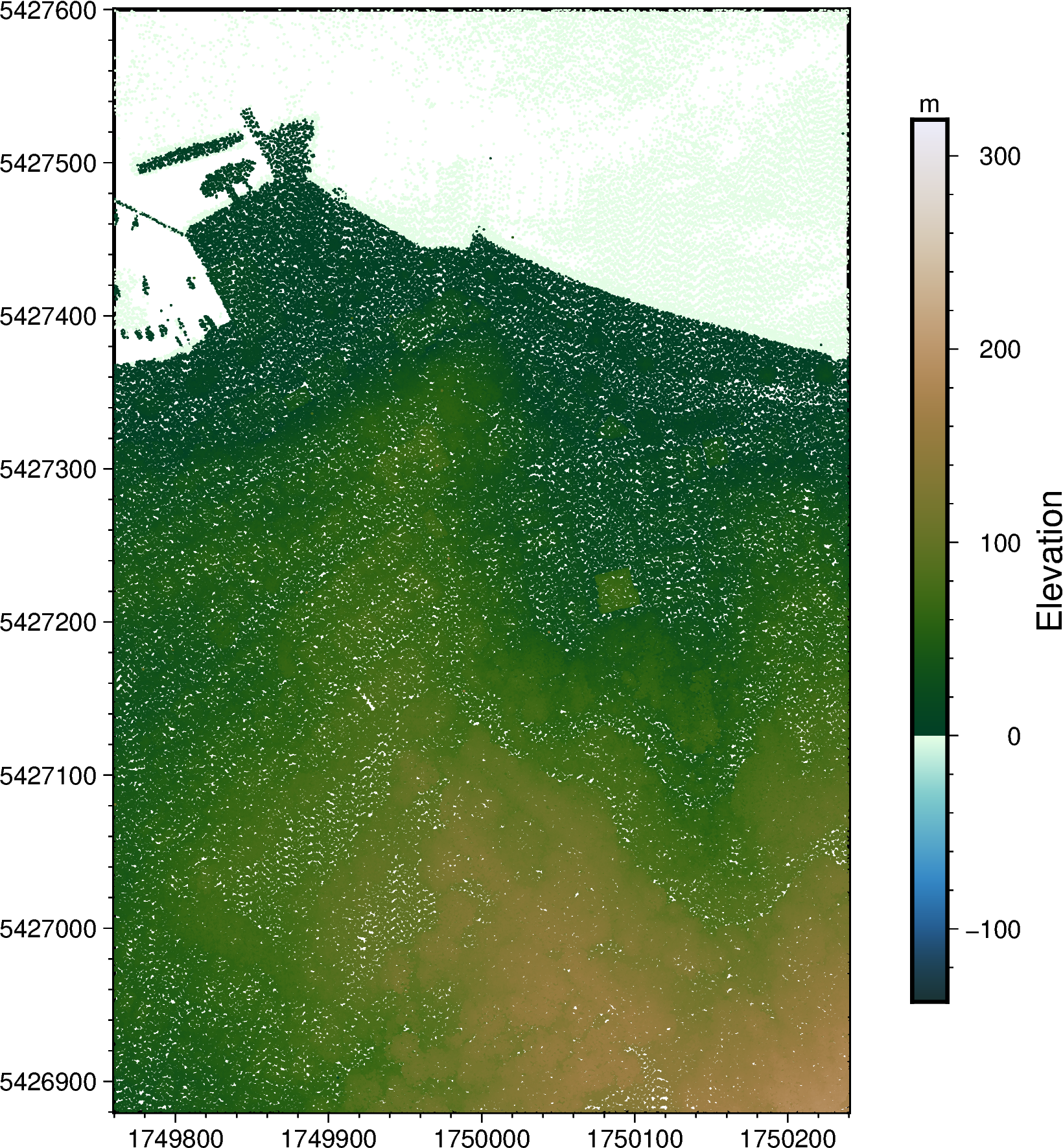
It’s hard to make out what the objects are, but hopefully you’ll see what looks like a wharf with ⛵ boats on the top left corner.
This is showing us a part of Oriental Bay in Wellington, with Mount Victoria ⛰️ towards the Southeast corner of the map.
1️⃣ Creating a Digital Surface Model (DSM)¶
Let’s now produce a digital surface model from our point cloud 🌧️
We’ll first do some proper spatial data cleaning 🧹 using both pandas and pygmt.
First step is to remove the LiDAR points classified as high noise 🔊 which is done by querying all the points that are not ‘18’.
🔖 References:
Table 17 of ASPRS LAS Specification
df = df.query(expr="classification != 18")
df
| x | y | z | classification | |
|---|---|---|---|---|
| 0 | 1749771.56 | 5427498.77 | -0.724 | 2 |
| 1 | 1749771.52 | 5427498.46 | -0.708 | 2 |
| 2 | 1749771.48 | 5427498.15 | -0.700 | 2 |
| 3 | 1749771.45 | 5427497.84 | -0.683 | 2 |
| 4 | 1749771.41 | 5427497.54 | -0.630 | 2 |
| ... | ... | ... | ... | ... |
| 7322687 | 1749760.95 | 5427567.17 | -0.479 | 9 |
| 7322698 | 1749760.23 | 5427567.65 | -0.488 | 9 |
| 7322699 | 1749760.26 | 5427567.98 | -0.442 | 9 |
| 7322700 | 1749760.29 | 5427568.29 | -0.417 | 9 |
| 7322701 | 1749760.32 | 5427568.62 | -0.404 | 9 |
7232453 rows × 4 columns
Get highest elevation points 🍫¶
Next, we’ll use the blockmedian function to trim 🪒 down the points spatially.
By default, this function computes a single median elevation for every pixel on an equally spaced grid 🏁
For a Digital Surface Elevation Model (DSM) though, we should pick the highest elevation 🔝 instead of the median.
So we’ll tell blockmedian to use the 99th quantile (T) instead, and set our output grid spacing to be exactly 1 metre (1+e) 📏
Note that we’ll only be taking the 🇽, 🇾, 🇿 (elevation) columns for this.
# Preprocess LiDAR data using blockmedian
df_trimmed = pygmt.blockmedian(
data=df[["x", "y", "z"]],
T=0.99, # 99th quantile, i.e. the highest point
spacing="1+e",
region=region,
)
df_trimmed
| x | y | z | |
|---|---|---|---|
| 0 | 1749760.05 | 5427599.54 | -0.755 |
| 1 | 1749760.67 | 5427599.83 | -0.737 |
| 2 | 1749761.87 | 5427599.71 | -0.747 |
| 3 | 1749765.39 | 5427599.55 | -0.701 |
| 4 | 1749765.96 | 5427599.73 | -0.742 |
| ... | ... | ... | ... |
| 311584 | 1750236.41 | 5426880.41 | 178.756 |
| 311585 | 1750237.32 | 5426880.47 | 179.132 |
| 311586 | 1750238.50 | 5426880.40 | 179.232 |
| 311587 | 1750239.36 | 5426880.46 | 179.517 |
| 311588 | 1750239.99 | 5426880.49 | 179.622 |
311589 rows × 3 columns
Turn points into a grid 🫓¶
Lastly, we’ll use pygmt.surface to produce a grid from the 🇽, 🇾, 🇿 data points.
Make sure to set our desired grid spacing to be exactly 📏 1 metre (1+e) again.
Also, we’ll set a tension factor (T) of 0.35 which is suitable for steep topography (since we have some 🏠 buildings with ‘steep’ vertical edges).
🚨 Note that this might take some time to run.
# Create a Digital Surface Elevation Model with
# a spatial resolution of 1m.
grid = pygmt.surface(
x=df_trimmed.x,
y=df_trimmed.y,
z=df_trimmed.z,
spacing="1+e",
region=region, # xmin, xmax, ymin, ymax
T=0.35, # tension factor
)
surface [WARNING]: 53246 unusable points were supplied; these will be ignored.
surface [WARNING]: You should have pre-processed the data with block-mean, -median, or -mode.
surface [WARNING]: Check that previous processing steps write results with enough decimals.
surface [WARNING]: Possibly some data were half-way between nodes and subject to IEEE 754 rounding.
Let’s take a look 👀 at the grid output.
The grid will be returned as an xarray.DataArray, with each pixel’s elevation (🇿 value in metres) stored at every 🇽 and 🇾 coordinate.
grid
<xarray.DataArray 'z' (y: 721, x: 481)>
array([[ 32.827084 , 33.058956 , 33.236935 , ..., 179.27199 ,
179.47954 , 179.68286 ],
[ 32.929073 , 33.571014 , 33.91027 , ..., 178.95854 ,
179.3416 , 179.49734 ],
[ 32.99532 , 34.62854 , 35.717705 , ..., 178.77904 ,
179.26633 , 179.20392 ],
...,
[ -0.7089864 , -0.7099118 , -0.7231264 , ..., -0.68712777,
-0.67993474, -0.65520626],
[ -0.7150018 , -0.72270995, -0.7585947 , ..., -0.6843457 ,
-0.68121314, -0.6624484 ],
[ -0.7903585 , -0.75000775, -0.78331774, ..., -0.70826954,
-0.68685687, -0.6937212 ]], dtype=float32)
Coordinates:
* x (x) float64 1.75e+06 1.75e+06 1.75e+06 ... 1.75e+06 1.75e+06
* y (y) float64 5.427e+06 5.427e+06 5.427e+06 ... 5.428e+06 5.428e+06
Attributes:
long_name: z
actual_range: [ -1.83348048 192.18907166]2️⃣ Plotting a Digital Surface Model (DSM)¶
Plot in 2D 🏳️🌈¶
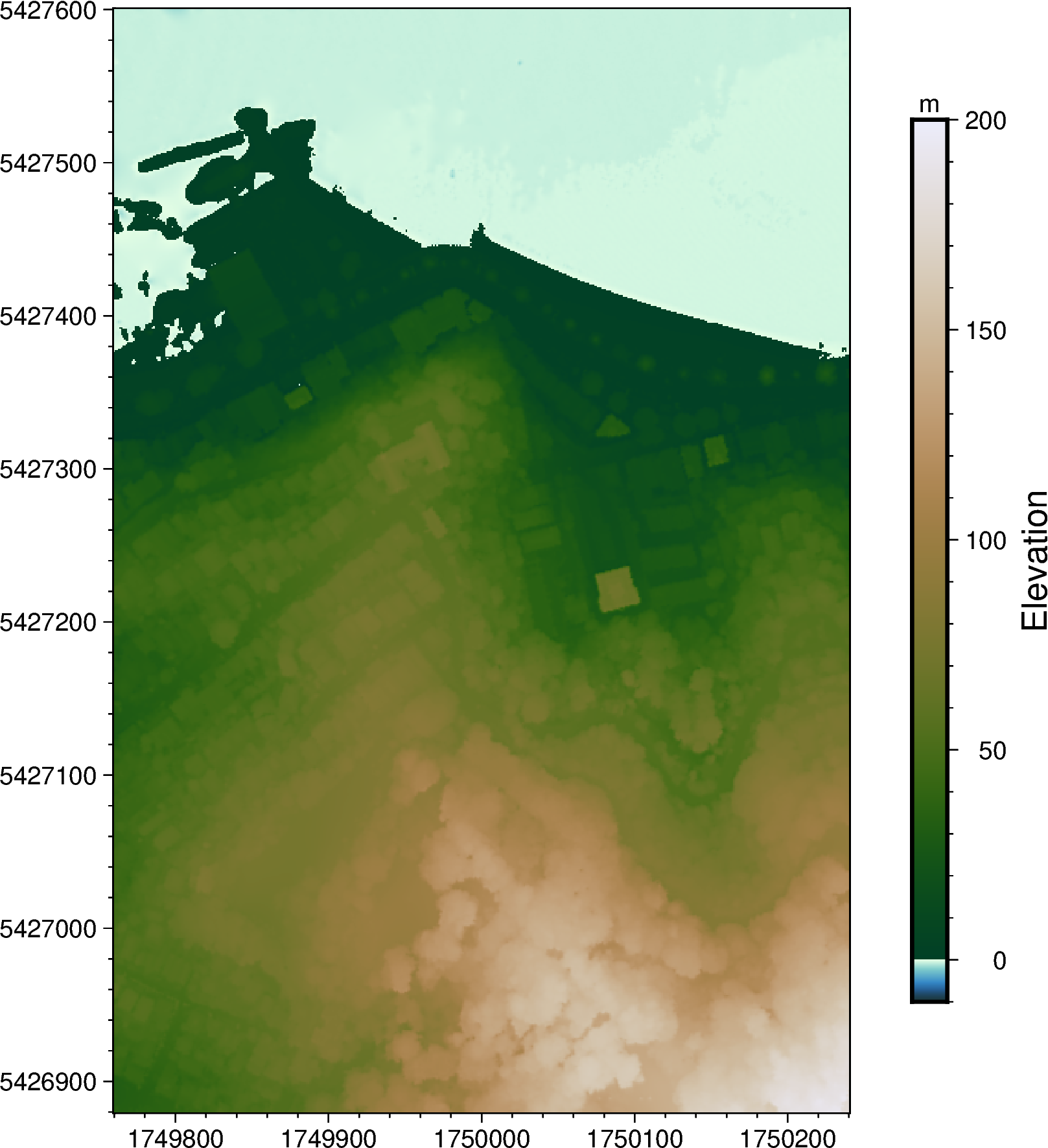
Now we can plot our Digital Surface Model (DSM) grid 🏁
This can be done by passing the xarray.DataArray grid into pygmt.Figure.grdimage.
fig2 = pygmt.Figure()
fig2.basemap(
frame=True,
region=[1_749_760, 1_750_240, 5_426_880, 5_427_600],
projection="x1:5000",
)
pygmt.makecpt(cmap="bukavu", series=[-10, 200])
fig2.grdimage(grid=grid, cmap=True)
fig2.colorbar(position="JMR", frame=["af+lElevation", "y+lm"])
fig2.show()
Plot 3D perspective view ⛰️¶
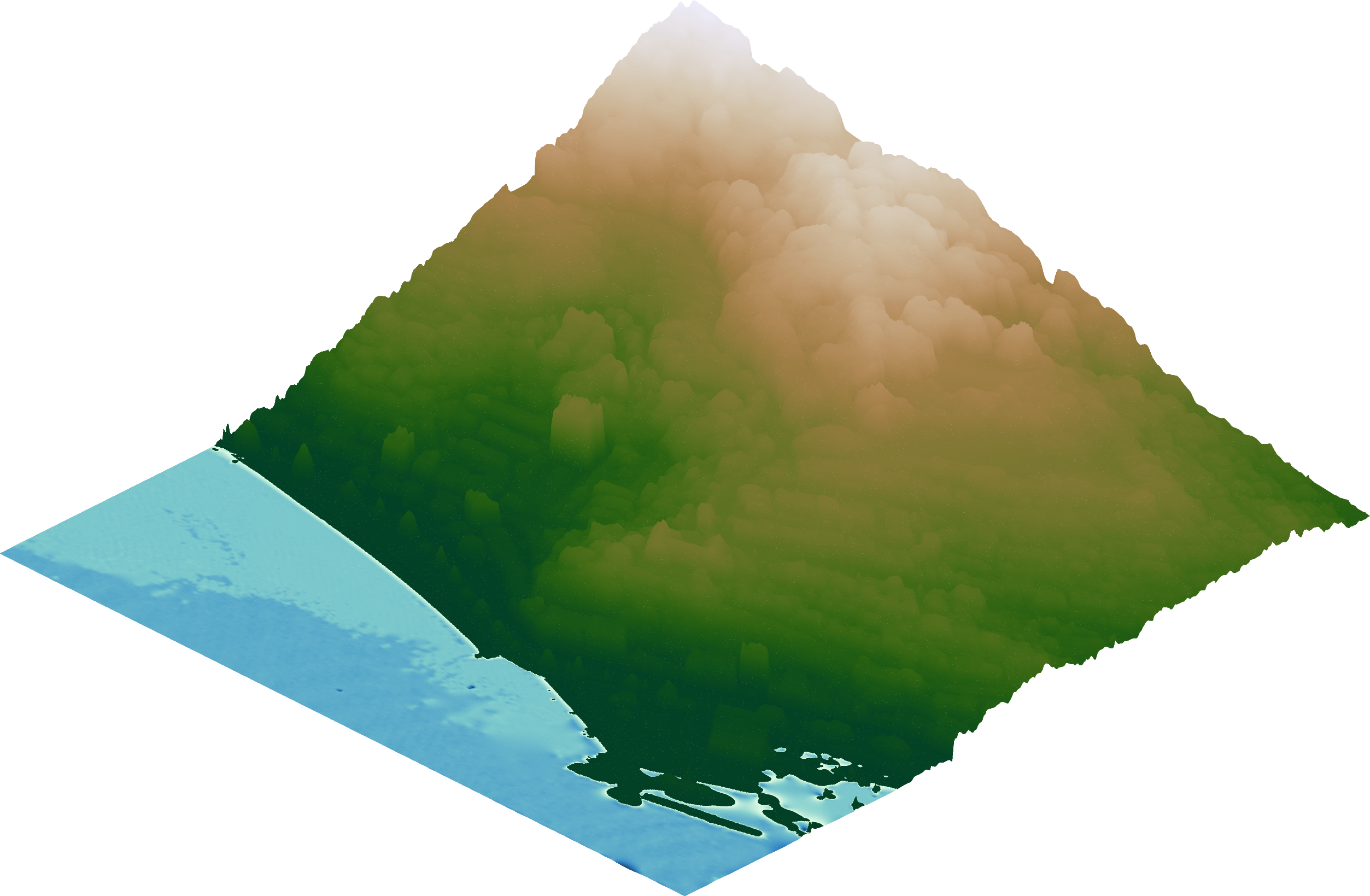
Now comes the cool part 💯
PyGMT has a grdview function to make high quality 3D perspective plots of our elevation surface!
Besides providing the elevation ‘grid’, there are a few basic things to configure:
cmap - name of 🌈 colormap to use
surftype - we’ll use ‘s’ here which just creates a regular surface 🏄
perspective - set azimuth angle (e.g. 315 for NorthWest) and 📐 elevation (e.g 30 degrees) of the viewpoint
zscale - a vertical exaggeration 🔺 factor, we’ll use 0.02 here
fig3 = pygmt.Figure()
fig3.grdview(
grid=grid,
cmap="bukavu",
surftype="s", # surface plot
perspective=[315, 30], # azimuth bearing, and elevation angle
zscale=0.02, # vertical exaggeration
)
fig3.show()
There are also other things we can 🧰 configure such as:
Hillshading ⛱️ using
shading=TrueA proper 3D map frame 🖼️ with:
automatic tick marks on x and y axis (e.g.
xaf+lLabel)z-axis automatic tick marks (
zaf+lLabel)plot title (
+tMy Title)
fig3 = pygmt.Figure()
fig3.grdview(
grid=grid,
cmap="bukavu",
surftype="s", # surface plot
perspective=[315, 30], # azimuth bearing, and elevation angle
zscale=0.02, # vertical exaggeration
shading=True, # hillshading
frame=[
"xaf+lEasting",
"yaf+lNorthing",
"zaf+lElevation (m)",
"+tOriental Bay, Wellington",
],
)
fig3.show()
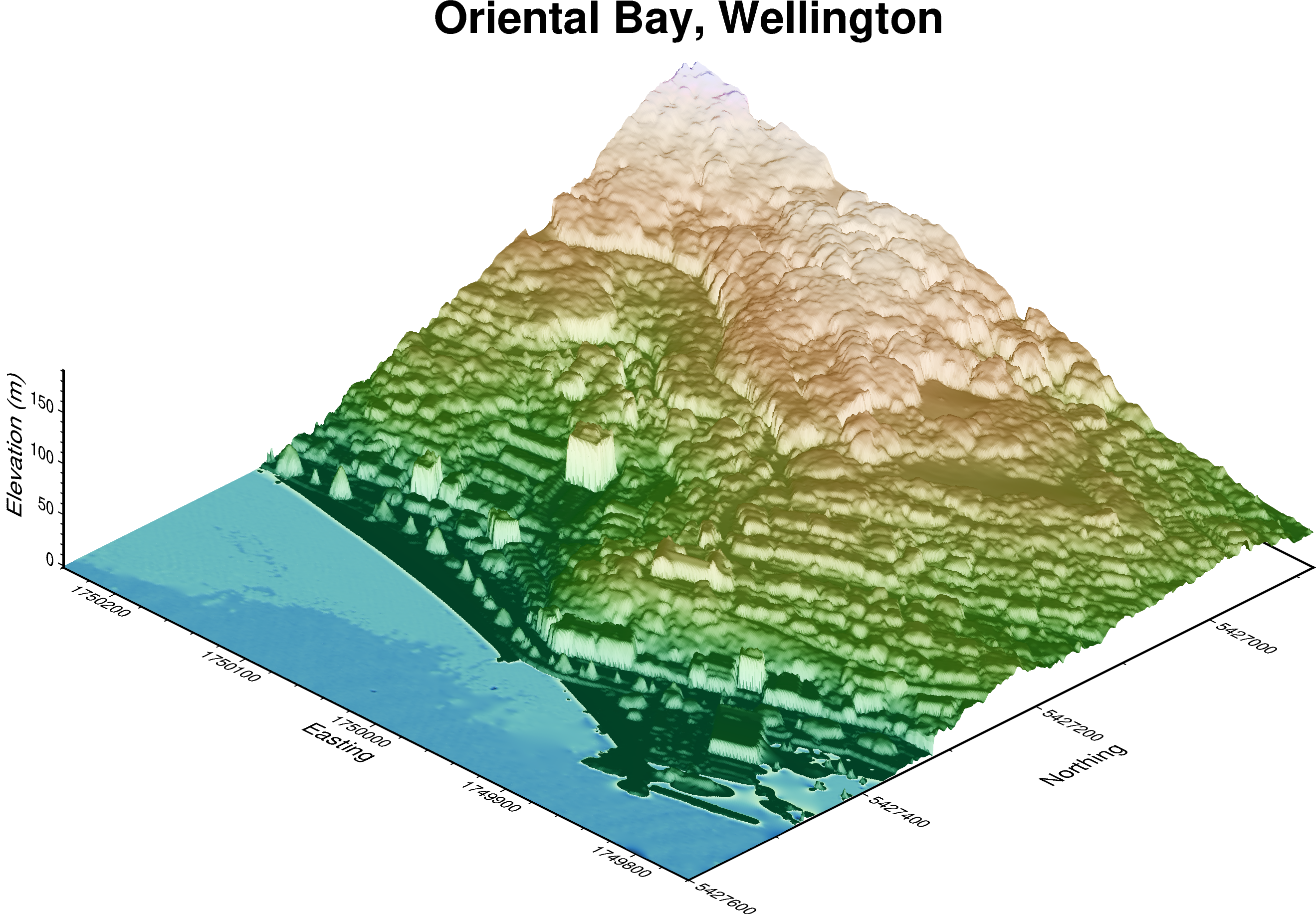
Feel free to have a go at changing the azimuth and elevation angle to get a different view 🔭
You can also try to tweak the vertical exaggeration factor 🗼 and play around with the map frame!
Save figures and output DSM to GeoTIFF 💾¶
Time to save your work!
We’ll save each of our figures into separate files first 🗃️
fig.savefig(fname="wellington_1d_lidar.png")
fig2.savefig(fname="wellington_2d_dsm_map.png")
fig3.savefig(fname="wellington_3d_dsm_view.png")
Also, it’ll be good to have a proper GeoTIFF of the DSM we made. This can be done using rioxarray’s to_raster method.
grid.rio.to_raster(raster_path="DSM_of_Wellington.tif")
The files should now appear on the file list on the left, and you can download them to your computer.
3️⃣ Bonus exercise - Create a 3D surface map of another area¶
Here’s a challenge to test your PyGMT skills 🤹
You’ll be processing a LiDAR point cloud of a different area 📍 from start to finish.
Good luck! 🥳
Feel free to find a dataset from an area you’re interested in using OpenTopography.
To make it easier though, I’ve provided an example for Queenstown, New Zealand 🇳🇿
OpenTopography link: https://doi.org/10.5069/G9MP51G0
Bulk download location: https://opentopography.s3.sdsc.edu/minio/pc-bulk/NZ21_Otago
Official 1m DSM from LINZ: https://data.linz.govt.nz/layer/105905-otago-queenstown-lidar-index-tiles-2021
These files covers the Central Queenstown area:
https://opentopography.s3.sdsc.edu/pc-bulk/NZ21_Otago/CL2_CC11_2021_1000_0813.laz
https://opentopography.s3.sdsc.edu/pc-bulk/NZ21_Otago/CL2_CC11_2021_1000_0814.laz
https://opentopography.s3.sdsc.edu/pc-bulk/NZ21_Otago/CL2_CC11_2021_1000_0913.laz
https://opentopography.s3.sdsc.edu/pc-bulk/NZ21_Otago/CL2_CC11_2021_1000_0914.laz
Download and load data¶
filenames = [
# "",
]
df = pd.DataFrame() # Start an empty DataFrame
for fname in filenames:
lazfile = pygmt.which(fname=fname, download=True)
lazdata = laspy.read(source=lazfile)
temp_df = pd.DataFrame(
data={
"x": lazdata.x.scaled_array(),
"y": lazdata.y.scaled_array(),
"z": lazdata.z.scaled_array(),
"classification": lazdata.classification,
}
)
# load each point cloud into the DataFrame
df = df.append(temp_df, ignore_index=True)
df
Plot the DSM¶
# Run grdimage
# Run grdview
That’s all 🎉! For more information on how to customize your map 🗺️, check out:
Tutorials at https://www.pygmt.org/v0.6.1/tutorials/index.html
Gallery examples at https://www.pygmt.org/v0.6.1/gallery/index.html
If you have any questions 🙋, feel free to visit the PyGMT forum at https://forum.generic-mapping-tools.org/c/questions/pygmt-q-a/11.
Submit any ✨ feature requests/bug reports to the GitHub repo at https://github.com/GenericMappingTools/pygmt
Cheers!